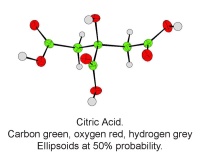
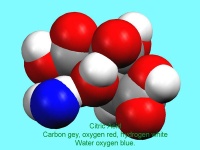
Figure 1: Andrew Cowley demonstrates the Nonius Kccd diffractometer to the Science Club. There is a second diffractometer behind the group
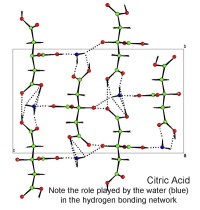
In October, Lynn Nickerson (Science Club Coordinator at Didcot Girls School) arranged for a small group to visit Chemical Crystallography in Oxford University’s new Chemistry Research Laboratory (Figure 1). The group was invited to bring some samples of common crystalline materials with them. The samples brought included cane sugar and citric acid (Figure 2). The girls used a polarising microscope to examine the crystals, and in the end selected an excellent crystal of citric acid for X-ray crystal structure determination. The crystal measured about 0.2 x 0.2 x 0.2 mm, and had to be ‘picked up’ on a fine nylon filament loop using a film of perfluoropolyether to hold it in place (Figure 3). The sample was put onto the Nonius kCCD automated diffractometer, cooled to -120°C and an X-ray diffraction image recorded (Figure 4). Dr Andrew Cowley collected a full data set in 40 minutes, which was processed by the Oxford crystallographic software CRYSTALS to reveal the structure of the acid (Figure 5 & 6). The hydrogen bonding network which holds the crystal together includes water of crystallisation, and is shown in Figure 7.

Figure 3: A single crystal of citric acid supported on a nylon loop. The ball point pen shows the scale